Car assembly lines use thousands of parts to build just one automobile, with each individual piece specifically engineered to perform one function. Could you instead build a car out of LEGO blocks, creating a complex machine out of a far more limited set of parts? This would require that each 'block' be very versatile: able to function in many different roles. However once in place, each block should adopt and retain only the role intended. This is the same strategy as that employed by life to maximize biochemical efficiency while also exploiting a dizzying array of chemical reactions. Nature's protein catalysis (called enzymes) accelerate reaction rates by up to 10,000,000,000,000,000,000-fold, yet do so using only 20 different amino acid building blocks and a dozen vitamin cofactors as additives. In studying enzyme catalysis, University of Kentucky graduate student Dongtao Cui and her professor Dr. Anne-Frances Miller hope to understand how these proteins can coerce their very versatile cofactors to perform only one reaction.
"Enzymes are large protein molecules that serve as nature's catalysis. They are built of strings amino acids folded and bunched up to create complex structures. Their ability to accelerate reactions is vital because the body only has sub-seconds to respond to a threat and must continuously replenish tissues and support organ function on a minute to minute basis” Miller said. Nonetheless reactions upon which we rely normally take days to eons at room temperature. Reactive cofactors bound by proteins execute these reactions in thousandths of a second. “The cofactor is a bit like a sparkplug. A motor does nothing for you without the sparkplugs. It’s heavy and useless until you put the sparkplugs in.” However what the cofactor does is as important as how fast it does it - like LEGO blocks, the functionality of a cofactor changes based on its location in the protein. “Different possible interactions between the protein and the cofactor explain why the same block (cofactor) does something different here than it did there,” Miller said. “Some sites can be modified, but the nature of this (cofactor) block can be modified by the (amino acid) blocks around it,” Cui added.
Cui obtained her bachelor’s degree from Beijing Normal University in 2001, and began her doctoral research with Miller at UK in 2005. “I chose Dr. Miller because I love instrument analysis and I feel that nuclear magnetic resonance (NMR) is so amazing. It’s a very powerful, very useful tool to investigate protein structures,” Cui said. While their research involves incredibly complex and subtle chemistry, the project undertaken by Cui and Miller has far-reaching, real world effects the go beyond the laboratory. Miller pointed out that understanding these proteins could potentially lead to a better understanding of syndromes such as MCAD (can cause death in a child whose blood sugar drops too low, and is associated with buildup of toxic metabolites). Children with MCAD have defective versions of enzymes that use a version of vitamin B2 to obtain energy from fats. The vitamin itself is fine (vitamin B2 is consumed as riboflavin and modified by our bodies for use). However the protein site at which it is bound can be defective. "If we knew what feature of the protein was responsible for the flavin’s ability to do chemistry, you would be in a position to devise fixes for it.” Other applications include using flavin-based enzymes to detoxify toxic waste. “Things like explosives which are still in the soil 50 or 60 years after the initial battles. Some of the enzymes we study are able to initiate their decomposition,” Miller explained. "Because of this extraordinary capability, it is even more important to understand the diverse functionality and use of these proteins. “We want to go in and change it, retaining the good behavior and eliminating the bad behavior.”
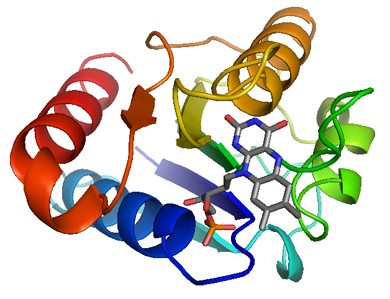
The ribbon diagram of flavodoxin. It is generated by interpolating a smooth curve through the polypeptide backbone. α-helices are shown as coiled ribbons, β-strands as arrows, and lines for random coils. The direction of the polypeptide chain is indicated by a color ramp along the length of the ribbon. The cofactor, FMN, is shown in stick model. Carbon, oxygen, nitrogen, and phosphorus atoms are colored in gray, red, blue, and orange, respectively.
